Abstract
Multiple Myeloma (MM) is a heterogeneous malignancy with genomic translocations and mutations conferring varying levels of risk of recurrence and confounding treatment options across patients. Intra-tumor clonal heterogeneity has also been observed which further complicates treatment. We hypothesize that the use of Single-Cell RNA sequencing (scRNA-seq) can elucidate the complex clonal transcriptomic structure of individual patients' disease, and can be used in concert with other genomic techniques to better characterize and treat patients on a personalized level.
Methods:
Individual CD138+ tumor cells were isolated and barcoded from bone marrow aspirates from eight relapsed myeloma patients using the 10x Genomics Chromium microfluidics platform and V2 chemistry for deep sequencing using an Illumina HiSeq 2500 machine. The 10x Genomics Chromium platform allows for large-scale simultaneous cellular sequencing which facilitates a more robust analysis of cellular populations than previous technologies. The number of cells analyzed ranged from 393 to 3,359 per sample, depending on sample cell suspension density. We used 10x Genomics Loupe software to visualize scRNA-seq data via TSNE dimensional reduction plots. Cells were then clustered via Graph-Based, K-Means, or custom gating methods.
Results:
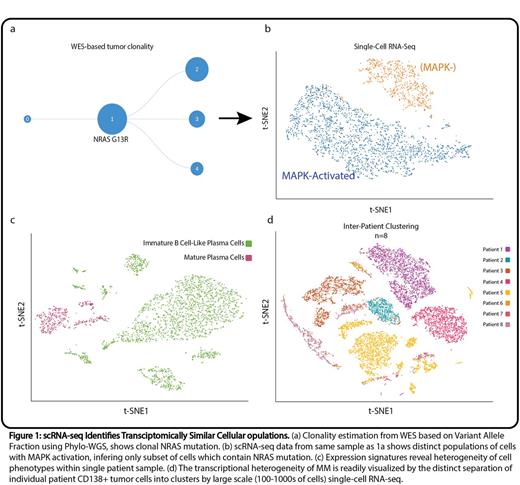
Analysis of bulk CD138+ whole-exome sequencing data has identified previously known mutational drivers and allowed estimation of cellular sub-populations (Fig. 1a) using copy-number alteration and mutational data via Phylo-WGS (Deshwar et al, Genome Biol 2015). Integrative analysis of bulk RNA-seq data revealed concordant activation of relevant pathways (e.g. MAP kinase) in patients with corresponding somatic mutations. On an individual patient level, we were able to process scRNA-seq data to visualize clusters of single cells with transcriptional similarity using t-distributed stochastic neighbor embedding (t-SNE) plots. We identified pathway activation in individual scRNA-seq clones via differential expression analysis and Signaling Pathway Impact Analysis (SPIA), in concordance with known somatic mutations (Fig. 1b). Genomic drivers and stages of differentiation in cell subsets (plasma cell vs immature B cell-like plasma cells) within a given patient were also revealed by distinct expression profiles (Fig. 1c). We utilized open-source drug repurposing tools with the RNA signature of individual clones to interrogate transcriptional signatures of drugs in public databases such as LINCS. The use of such drug repurposing methods will be effective in generating more targeted personalized therapies, utilizing cluster specific expression profiles. This is especially useful for cellular populations which do not contain a known target based on DNA mutational analysis. In a pooled analysis of all samples, each patient sample remarkably formed its own spatially distinct cluster of cells (Fig. 1d). These data suggest that relapsed myeloma patients form transcriptionally distinct clones, and myeloma therapy may need to be highly individualized to overcome clonal heterogeneity. scRNA-seq is a powerful tool which can be utilized alone, or in harmony with other sequencing technologies and genomic techniques, to improve characterization and treatment of myeloma patients in a clone specific manner.
Madduri: Foundation Medicine, Inc.: Consultancy. Chari: Novartis: Consultancy, Research Funding; Celgene Corporation: Consultancy, Research Funding; Millennium Pharmaceuticals, Inc.: Consultancy, Research Funding; Janssen: Consultancy, Research Funding; Array BioPharma: Consultancy, Research Funding; Amgen: Honoraria, Research Funding. Cho: Genentech: Other: advisory board, Research Funding; Bristol Myers-Squibb: Other: advisory board, Research Funding; Ludwig Institute for Cancer Research: Research Funding; Multiple Myeloma Research Foundation: Research Funding; Agenus, Inc.: Research Funding. Barlogie: Celgene Corporation: Consultancy, Research Funding; Millenium Pharmaceuticals: Consultancy, Research Funding. Jagannath: MMRF: Speakers Bureau; Medicom: Speakers Bureau; Merck: Consultancy; Novartis: Consultancy; Celgene: Consultancy; Bristol-Meyers Squibb: Consultancy. Dudley: NuMedii, Inc.: Equity Ownership, Patents & Royalties; Janssen Pharmaceuticals, Inc.: Consultancy; Ayasdi, Inc.: Equity Ownership; Ecoeos, Inc.: Equity Ownership; Ontomics, Inc.: Equity Ownership; Personalis: Patents & Royalties; AstraZeneca: Consultancy; GlaxoSmithKline: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal